Research Focus
To carry out their biological functions, macromolecules are often modified by small chemical tags. These so called post-translational and post-transcriptional modifications have profound impact on function by affecting the ability of macromolecules to localize in cellular compartments, interact with their partners or small molecule drugs, or propagate signals, among other roles. Our research program investigates mechanisms that regulate modifications, studies consequences of modifications in physiology and disease pathogenesis, and develops chemical probes to perturb modifications. We are particularly interested in methylation of two essential macromolecular assemblies – the chromatin and the ribosome.
|
Methylation in Epigenetics
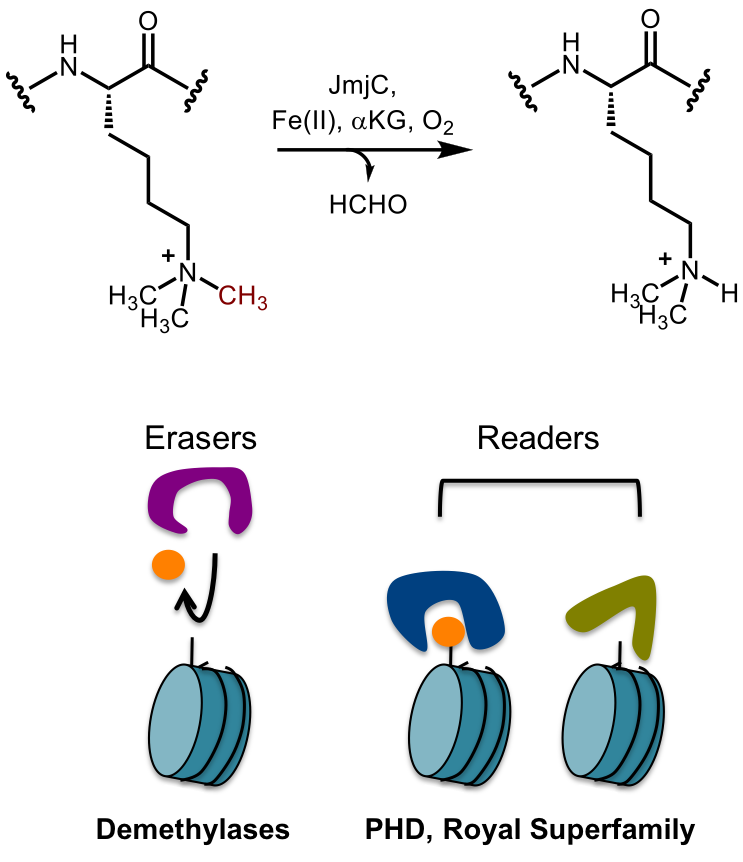
Chemical modifications of chromatin are critical regulators of chromatin structure and gene expression. To define mechanisms that regulate chromatin methylation status, we investigate catalysis carried out by histone demethylases, a class of epigenetic ‘eraser’ proteins that antagonize chromatin methylation. We investigate functional coupling between the catalytic domain and chromatin reader domains in these enzymes, with goals of identifying chromatin-mediated regulatory mechanisms and uncovering potential druggable sites. |
|
Chemical Probes for Chromatin Methylation
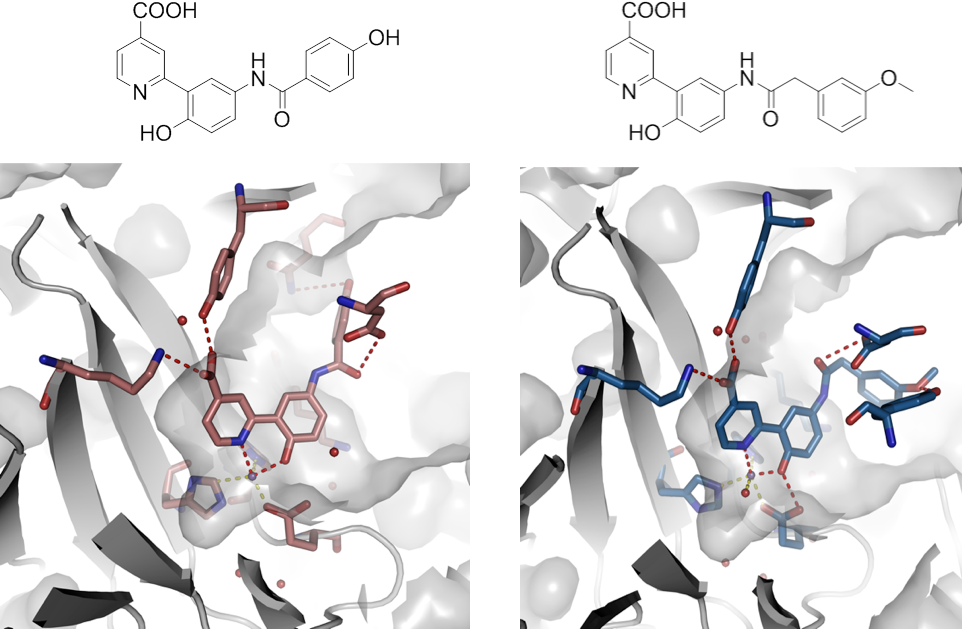
Our group is developing chemical tool compounds to interrogate erasers and readers of chromatin methylation. These molecules can enable target validation studies on proteins that recognize and remove methylation to facilitate transcriptional regulation in cancer. Inhibitors that target allosteric and orthosteric sites, as well as degraders, are being investigated.
|
|
Methylation in Antibiotic Resistance
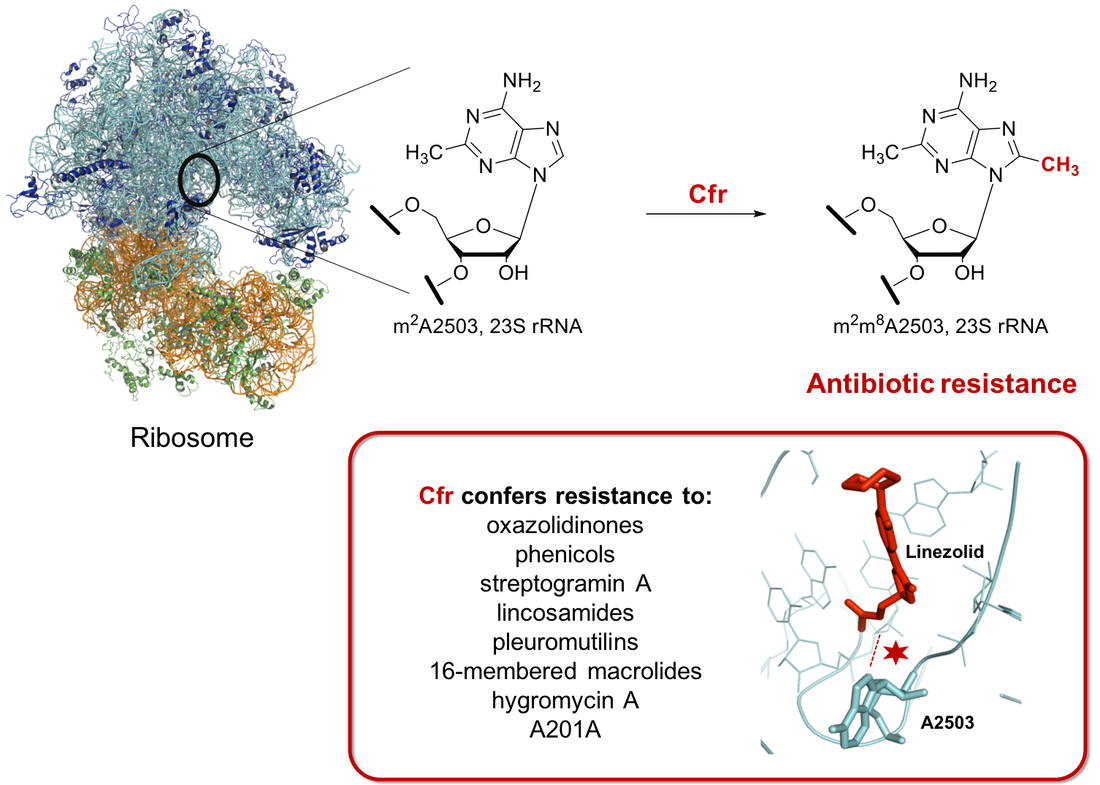
Chloramphenicol-florfenicol resistance protein, Cfr, confers resistance to a number of antibiotics, including linezolid. Cfr is a methylating enzyme that modifies the bacterial ribosome. We investigate mechanism of the unusual methylation catalyzed by this enzyme, as well as the molecular basis of antibiotic resistance conferred by the methylation. In addition, we are developing and utilizing transcriptome-wide techniques to de-orphan RNA methylating enzymes. |

Proudly powered by Weebly