Publications
|
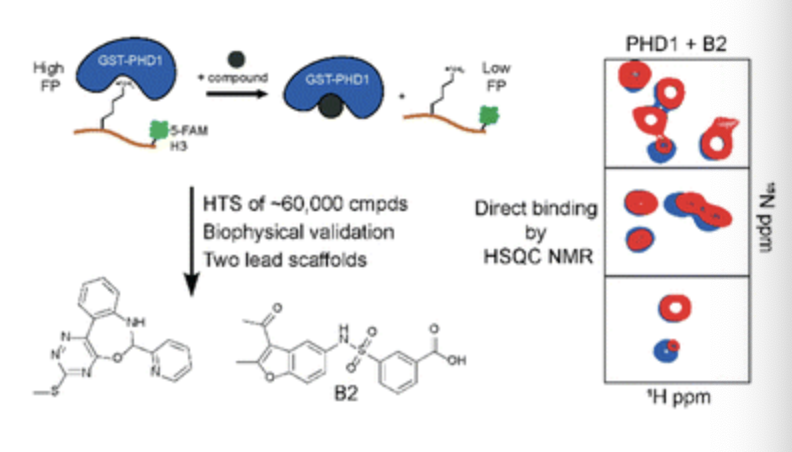
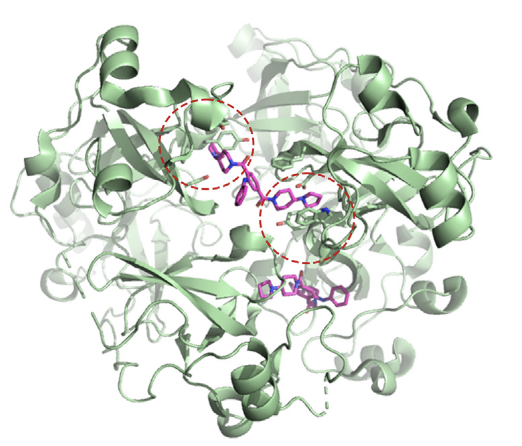
Ortiz G, Longbotham JE, Qin SL, Zhang MY, Lee GM, Neitz RJ, Kelly MJS, Arkin MR, Fujimori DG. Identifying Ligands for the PHD1 Finger of KDM5A Through High-Throughput Screening. RSC Chem. Bio. 2023 December 18.
Full Text |
|
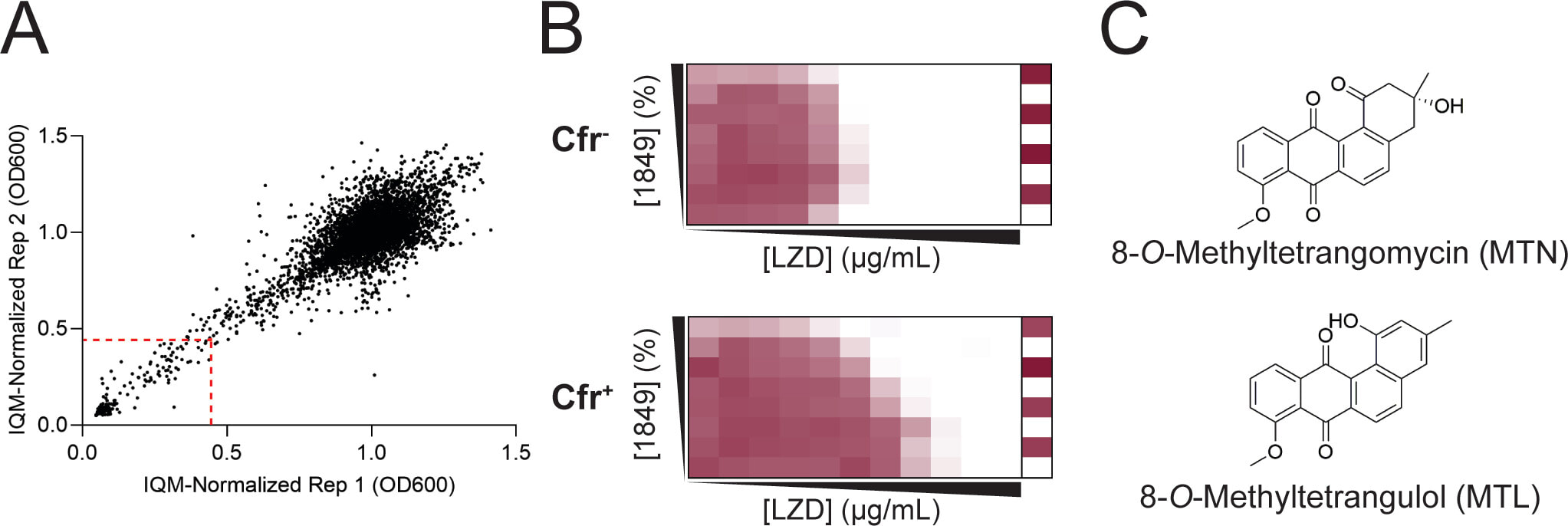
Schaenzer AJ, Rodriguez-Hernandez A, Tsai K, Hobson C, Fujimori DG, Wright GD. Angucylinones Rescue PhLOPSA Antibiotic Activity by Inhibiting Cfr-Dependent Antibiotic Resistance. mBio. 2023 November 28.
Full Text |
|
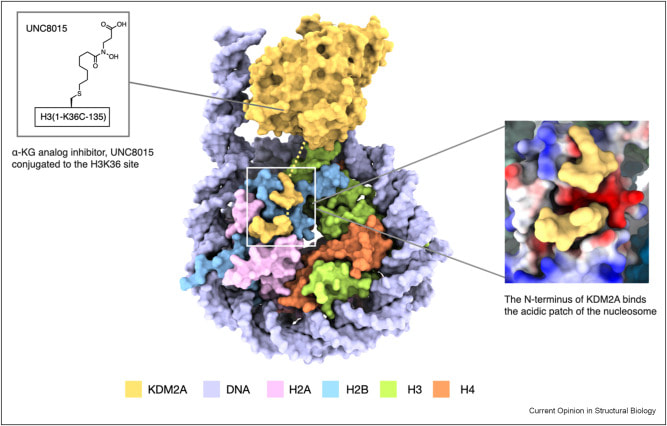
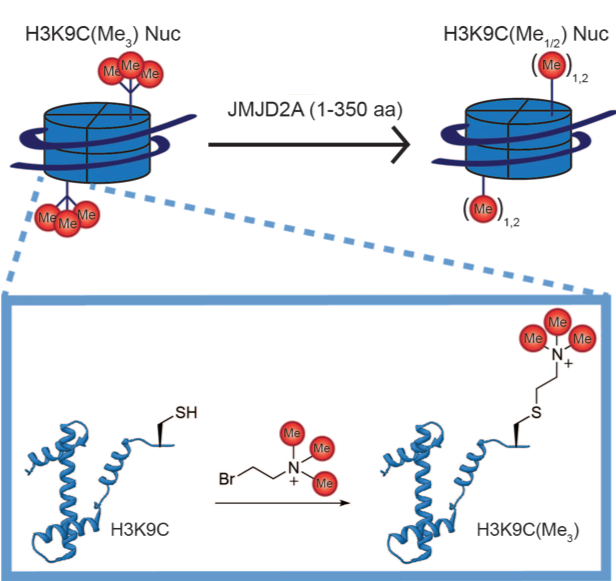
Sarah L, Fujimori DG. Recent Developments in Catalysis and Inhibition of the Jumonji Histone Demethylases. Curr. Opin. Struct. Bio. 2023 October 11.
Full Text |
|
|
|
|
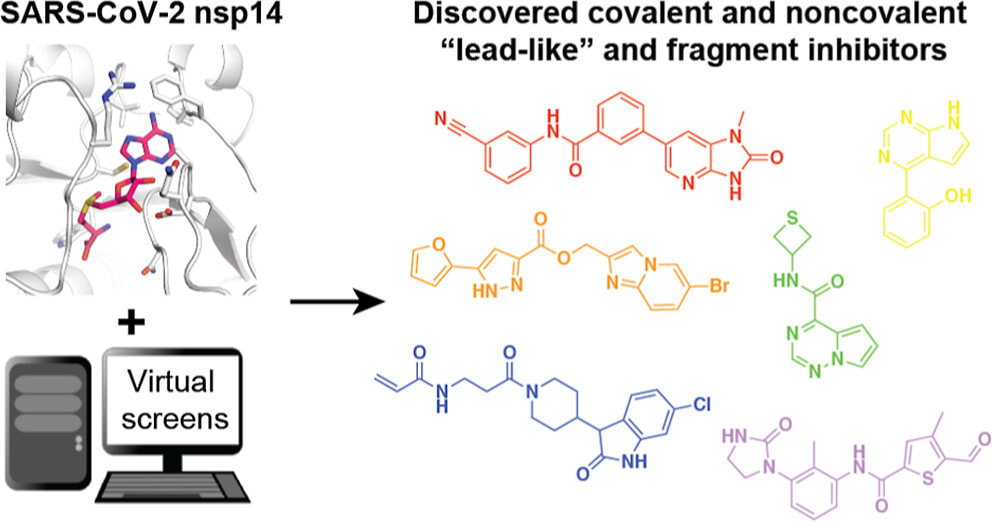
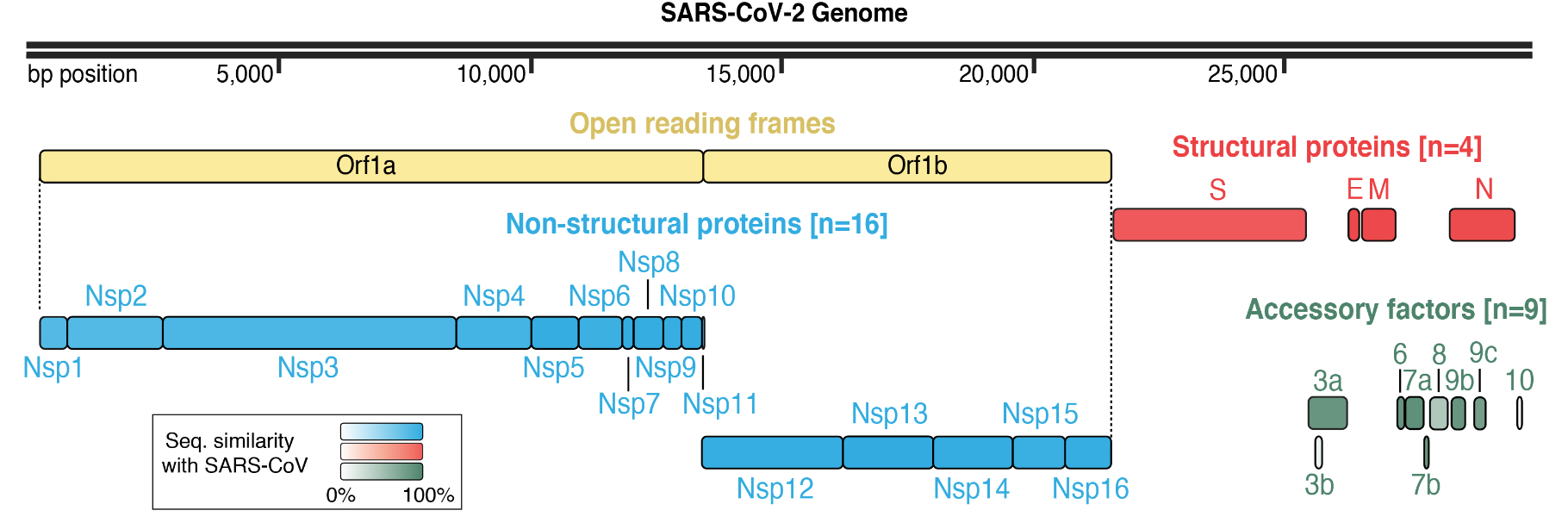
Singh I, Li F, Fink EA, Chau I, Li A, Rodriguez-Hernandez A, Glenn I, Zapatero-Belinchón FJ, Rodriguez ML, Devkota K, Deng Z, White K, Wan X, Tolmachova NA, Moroz YS, Kaniskan HÜ, Ott M, García-Sastre A, Jin J, Fujimori DG, Irwin JJ, Vedadi M, Shoichet BK. Structure-based discovery of inhibitors of the SARS-CoV-2 Nsp14 N7-methyltransferase. J. Med. Chem. 2023 June 9.
Full text |
|
Ortiz G, Kutateladze TG, Fujimori DG. Chemical tools targeting readers of lysine methylation. Curr Opin Chem Biol. 2023 January.
Full text |
|
Ugur FS, Kelly MJS, Fujimori DG. Chromatin Sensing by the Auxiliary Domains of KDM5C Regulates Its Demethylase Activity and Is Disrupted by X-linked Intellectual Disability Mutations. J. Mol. Biol. 2023 January 30.
Full text |
|
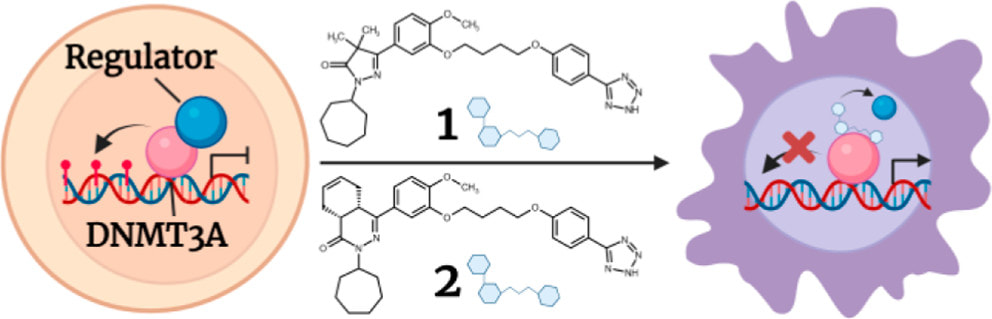
Sandoval JE, Ramabadran R, Stillson N, Sarah L, Fujimori DG, Goodell MA, Reich N. First-in-Class Allosteric Inhibitors of DNMT3A Disrupt Protein-Protein Interactions and Induce Acute Myeloid Leukemia Cell Differentiation. J. Med. Chem. 2022 July 22.
Full text |
|
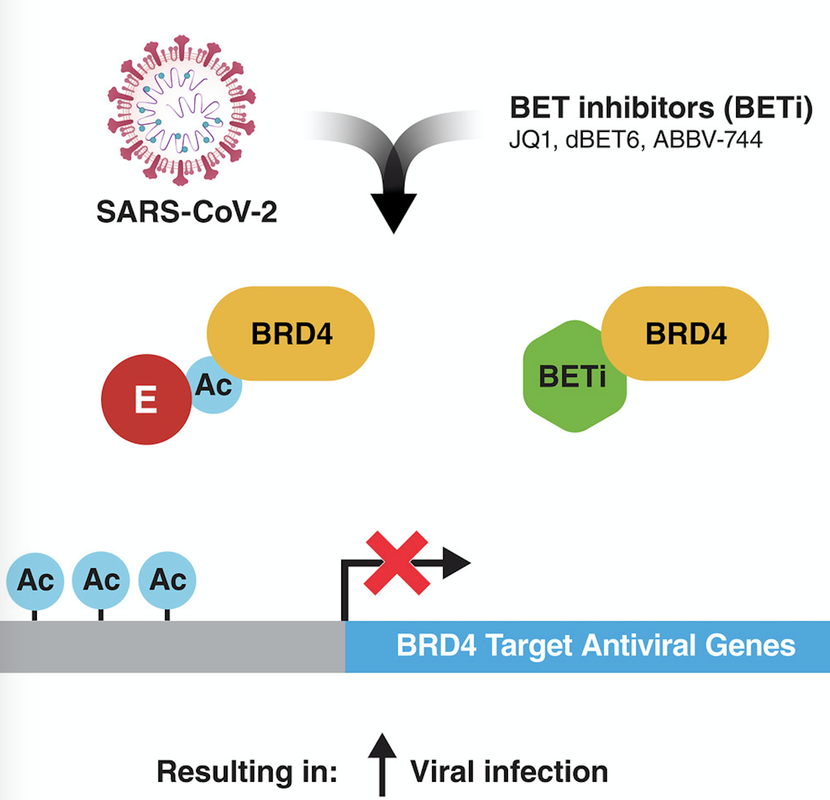
Chen IP, Longbotham JE, McMahon S, Suryawanshi RK, Khalid MM, Taha TY, Tabata T, Hayashi JM, Soveg FW, Carlson-Stevermer J, Gupta M, Zhang MY, Lam VL, Li Y, Yu Z, Titus EW, Diallo A, Oki J, Holden K, Krogan N, Fujimori DG, Ott M. Viral E Protein Neutralizes BET Protein-Mediated Post-Entry Antagonism of SARS-CoV-2. Cell Rep. 2022 June 27.
Full text |
|
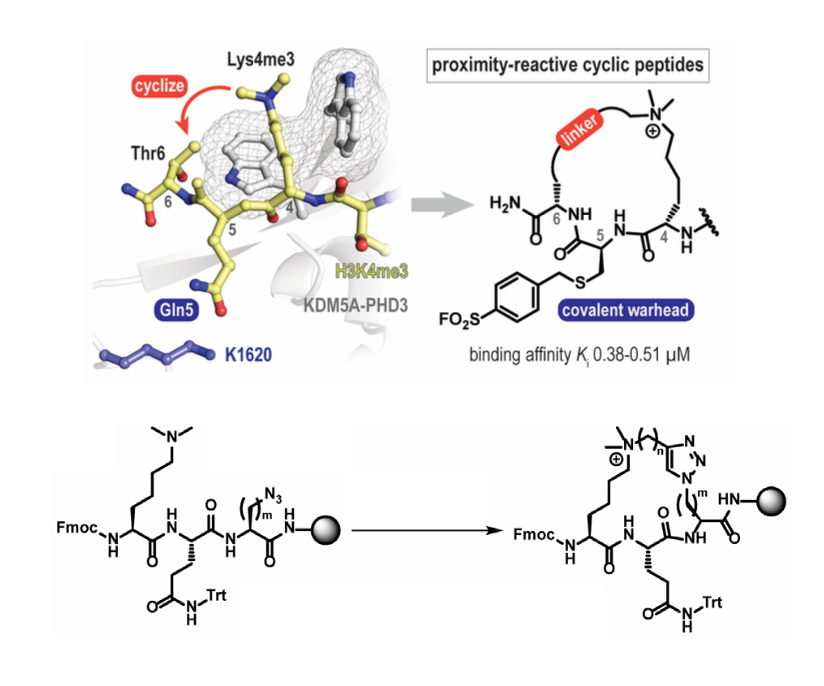
Zhang MY, Yang H, Ortiz G, Trnka MJ, Petronikolou N, Burlingame AL, DeGrado B, Fujimori DG. Covalent labeling of a chromatin reader domain using proximity- reactive cyclic peptides. Chem. Sci. 2022 May 10.
Full text |
|
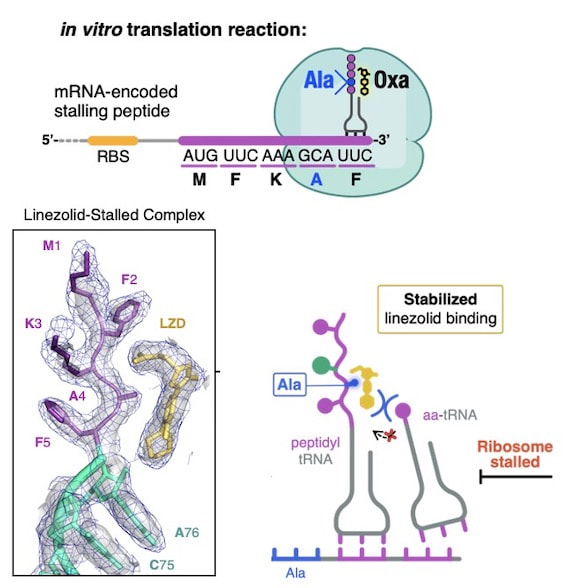
Tsai K, Stojković V, Lee DJ, Young ID, Szal T, Vasquez-Laslop N, Mankin AS, Fraser JS, Fujimori DG. Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat. Struct. Mol. Biol. 2022 Feb 29. Full Text |
|
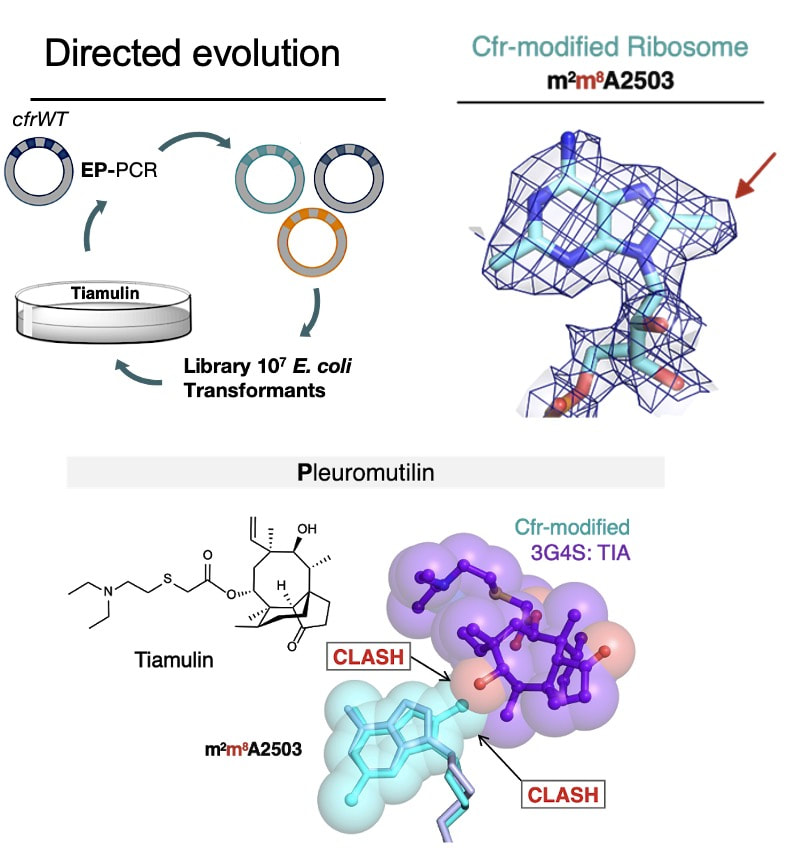
Tsai K, Stojković V, Noda-Garcia L, Young ID, Myasnikov AG, Kleinman J, Palla A, Floor SN, Frost A, Fraser JS, Tawfik DS, Fujimori DG. Directed evolution of the rRNA methylating enzyme Cfr reveals molecular basis of antibiotic resistance. eLife 2022 Jan 11.
Full Text |
|
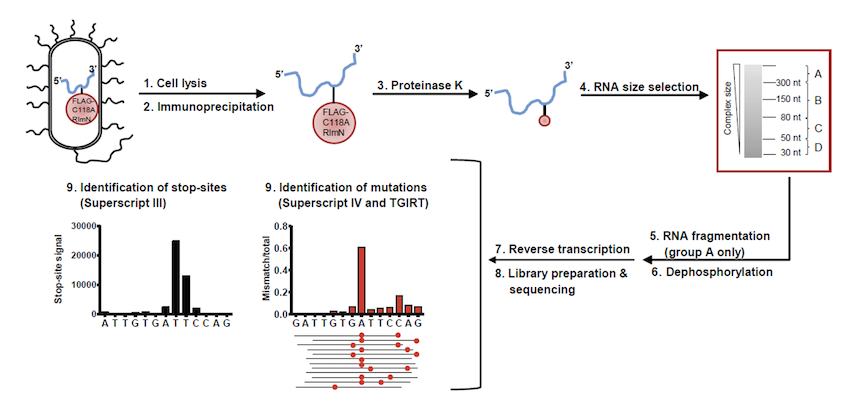
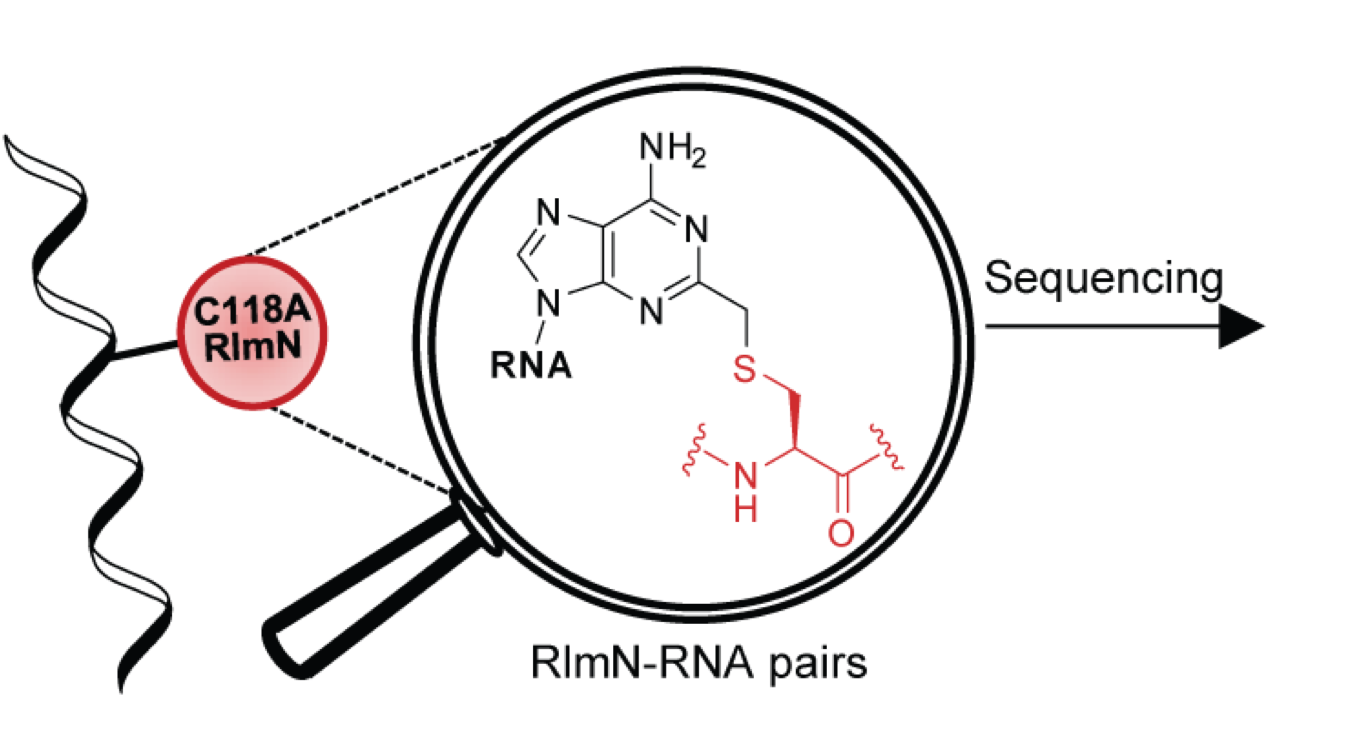
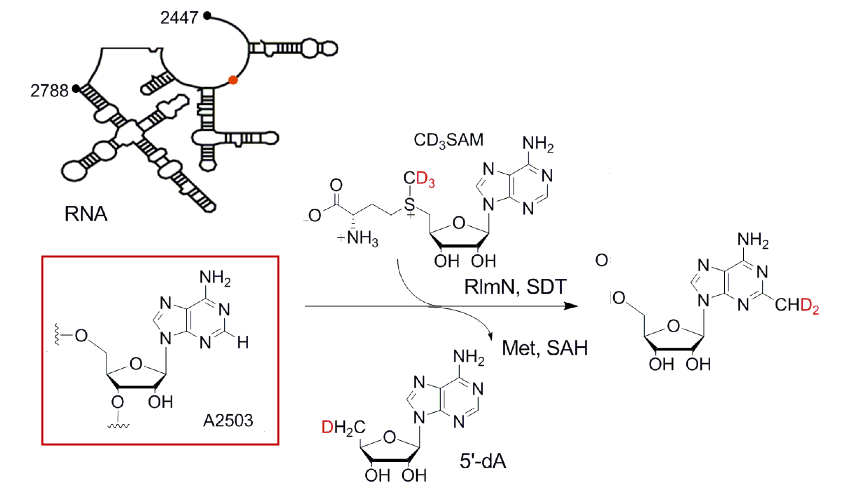
Stojković V, Weinberg DE, Fujimori DG. miCLIP-MaPseq Identifies Substrates of Radical SAM RNA-Methylating Enzyme Using Mechanistic Cross-Linking and Mismatch Profiling. Methods Mol. Biol. 2021 Jun 4.
Full Text |
|
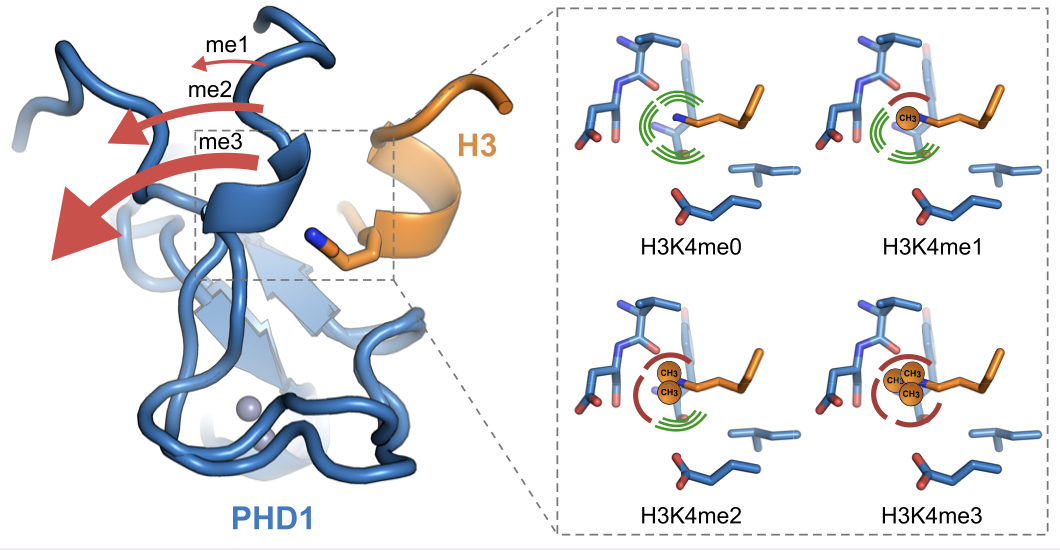
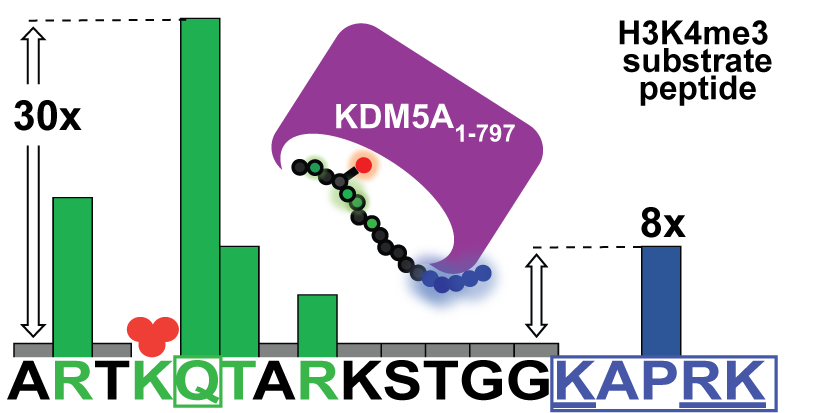
Longbotham JE, Kelly MJS, Fujimori DG. Recognition of Histone H3 Methylation States by the PHD1 Domain of Histone Demethylase KDM5A. ACS Chem. Biol. 2021 Feb 18.
Full Text |
|
Anderson SE, Longbotham JE, O'Kane PT, Ugur FS, Fujimori DG, Mrksich M. Exploring the Ligand Preferences of the PHD1 Domain of Histone Demethylase KDM5A Reveals Tolerance for Modifications of the Q5 Residue of Histone 3. ACS Chem. Biol. 2020 Dec 14.
Full Text |
|
Longbotham JE, Zhang ME, Fujimori DG. Domain cross-talk in regulation of histone modifications: Molecular mechanisms and targeting opportunities. Curr Opin Chem Biol 2020 Aug 2020;57:105-113
Full Text |
|
Petronikolou N, Longbotham JE, Fujimori DG. Dissecting contributions of catalytic and reader domains in regulation of histone demethylation. Methods Enzymol 2020 Apr, 639:217-236 Full Text |
|
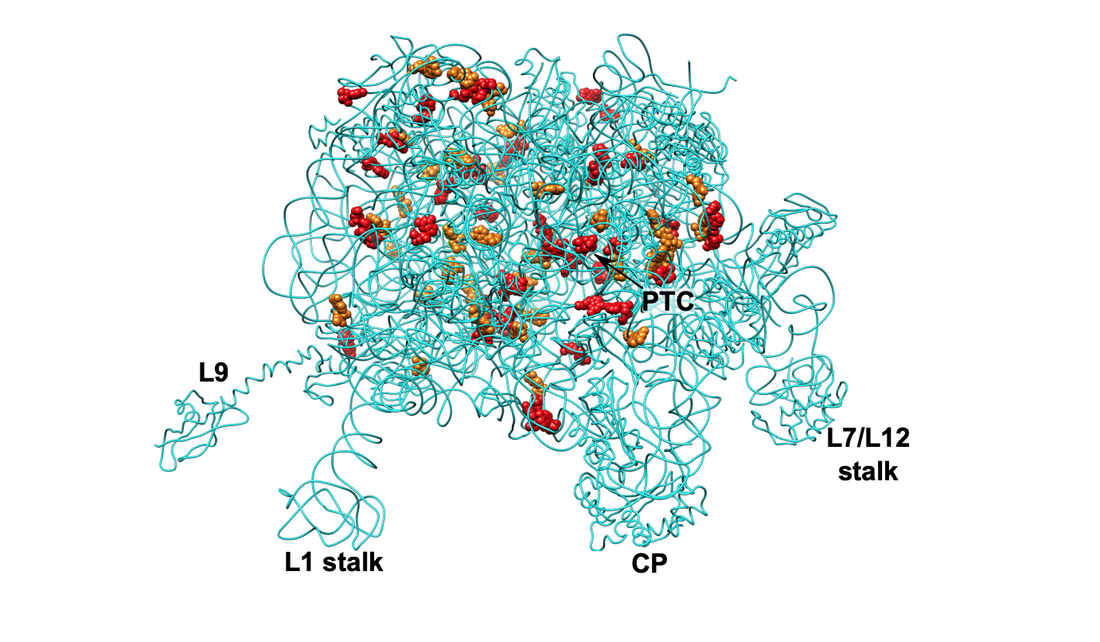
Stojković V, Myasnikov AG, Young ID, Frost A, Fraser JS, Fujimori DG. Assessment of the Nucleotide Modifications in the High-Resolution Cryo-Electron Microscopy Structure of the Escherichia Coli 50S Subunit. Nucleic Acids Res 2020 Mar 18;48(5):2723-2732 PMID: 31989172 Deposited Structure: 6PJ6 Full Text |
|
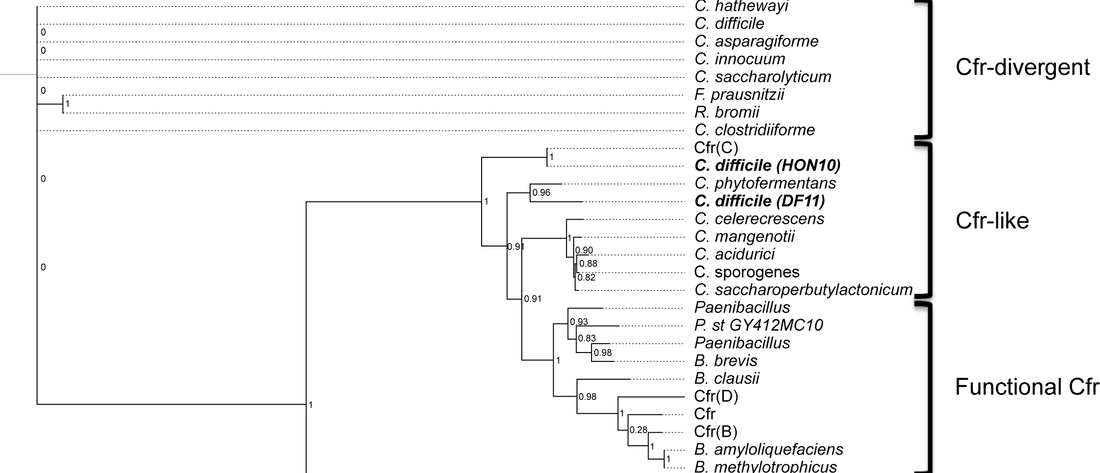
Stojković V, Ulate ME, Hidalgo F, Aguilar E, Monge-Cascante C,Pizarro M, Tsai K, Tzoc E, Camorlinga M, Paredes-Sabja D, Quesada-Gómez C, Fujimori DG, Rodríguez C. cfr(B), cfr(C), and a new cfr-like gene, cfr(E), in Clostridium difficile strains recovered across Latin America. Antimicrob. Agents Chemother 2019 Nov 4, AAC.01074-19
PMID: 31685464 Full Text |
|
Stojković V, Chu T, Therizols G, Weinberg DE, Fujimori DG. miCLIP-MaPseq, a Substrate Identification Approach for Radical SAM RNA Methylating Enzymes. J Am Chem Soc 2018 Jun 13; 140(23): 7135-7143 PMID: 29782154 Full Text |
|
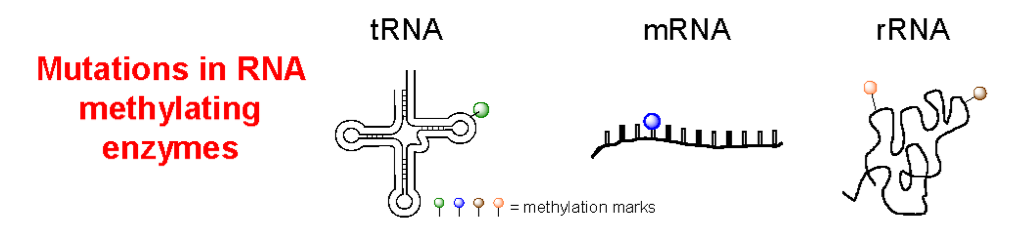
Stojković V, Fujimori DG. Mutations in RNA methylating enzymes in disease. Curr Opin Chem Biol 2017 Dec; 41:20-27 (review) PMCID: PMC5723530 Full Text |
|
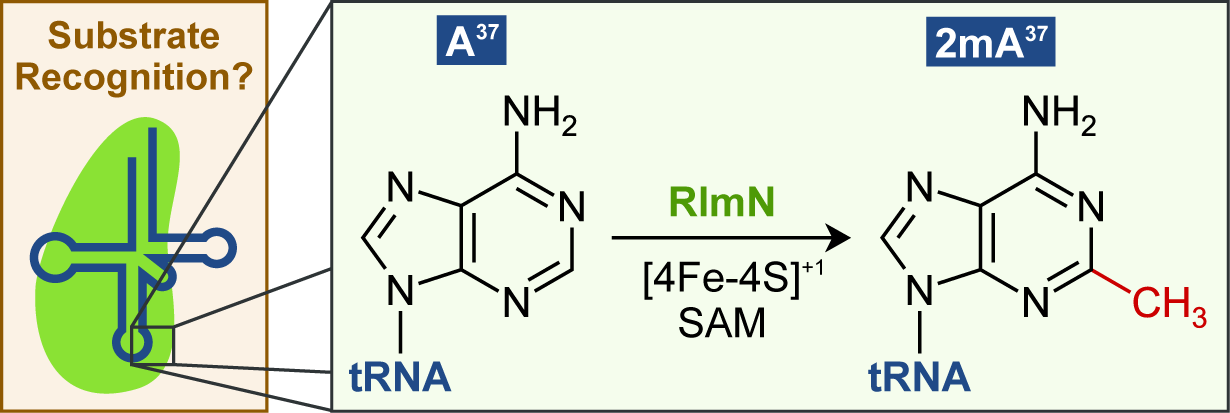
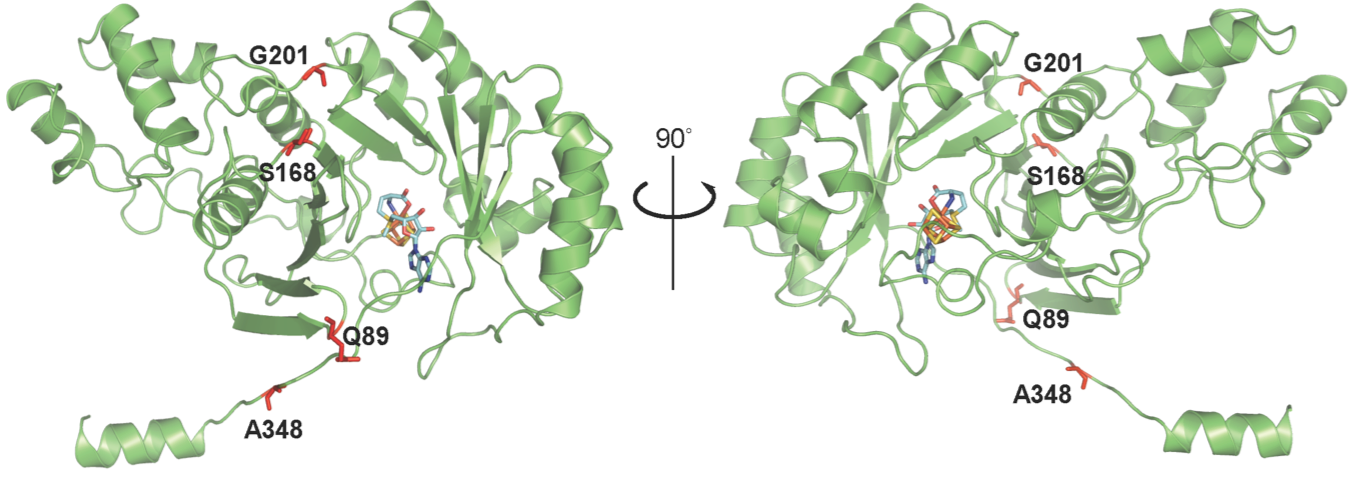
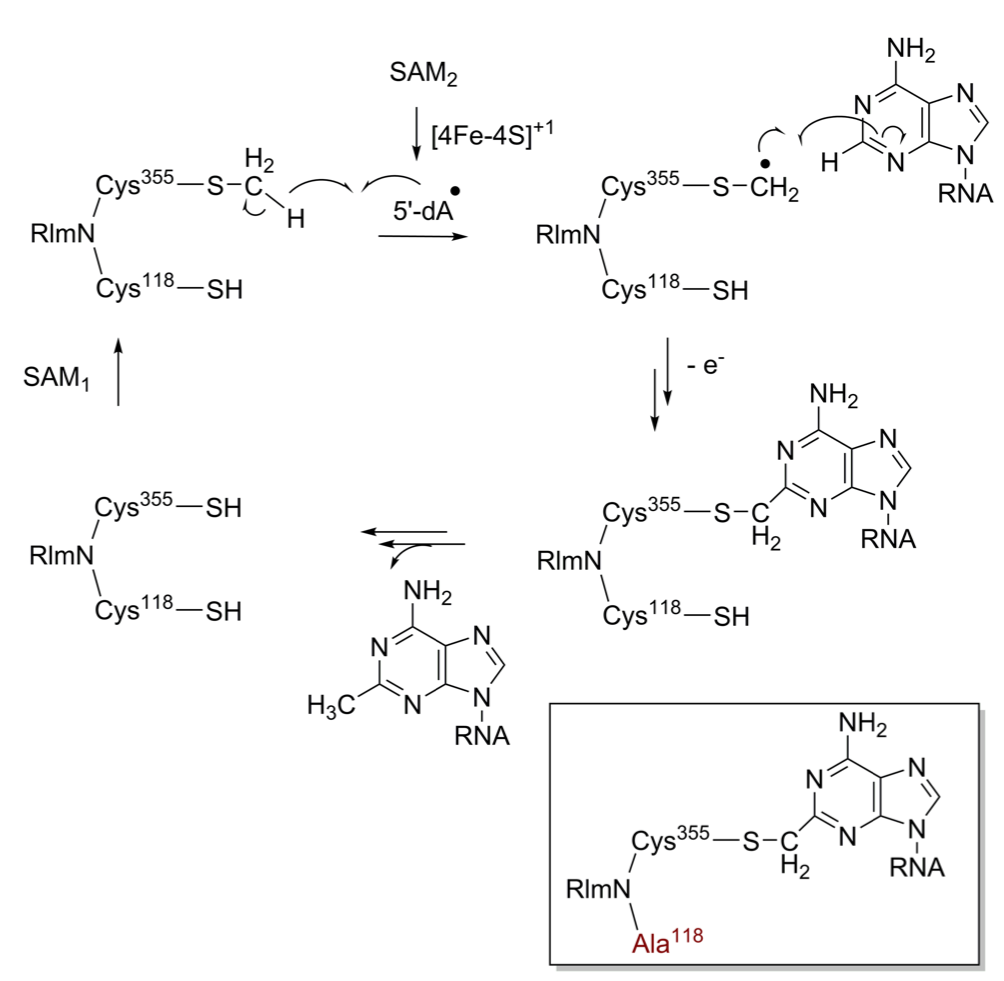
Fitzsimmons CM, Fujimori DG. Determinants of tRNA Recognition by the Radical SAM Enzyme RlmN. Plos One 2016 Nov 30;11(11):e0167298 PMCID: PMC5130265 Full Text |
|
Stojković V, Noda-Garcia L, Tawfik DS, Fujimori DG. Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme. Nucleic Acids Res 2016 Oct 14;44(18):8897-8907 PMCID: PMC5062987 Full Text |
|
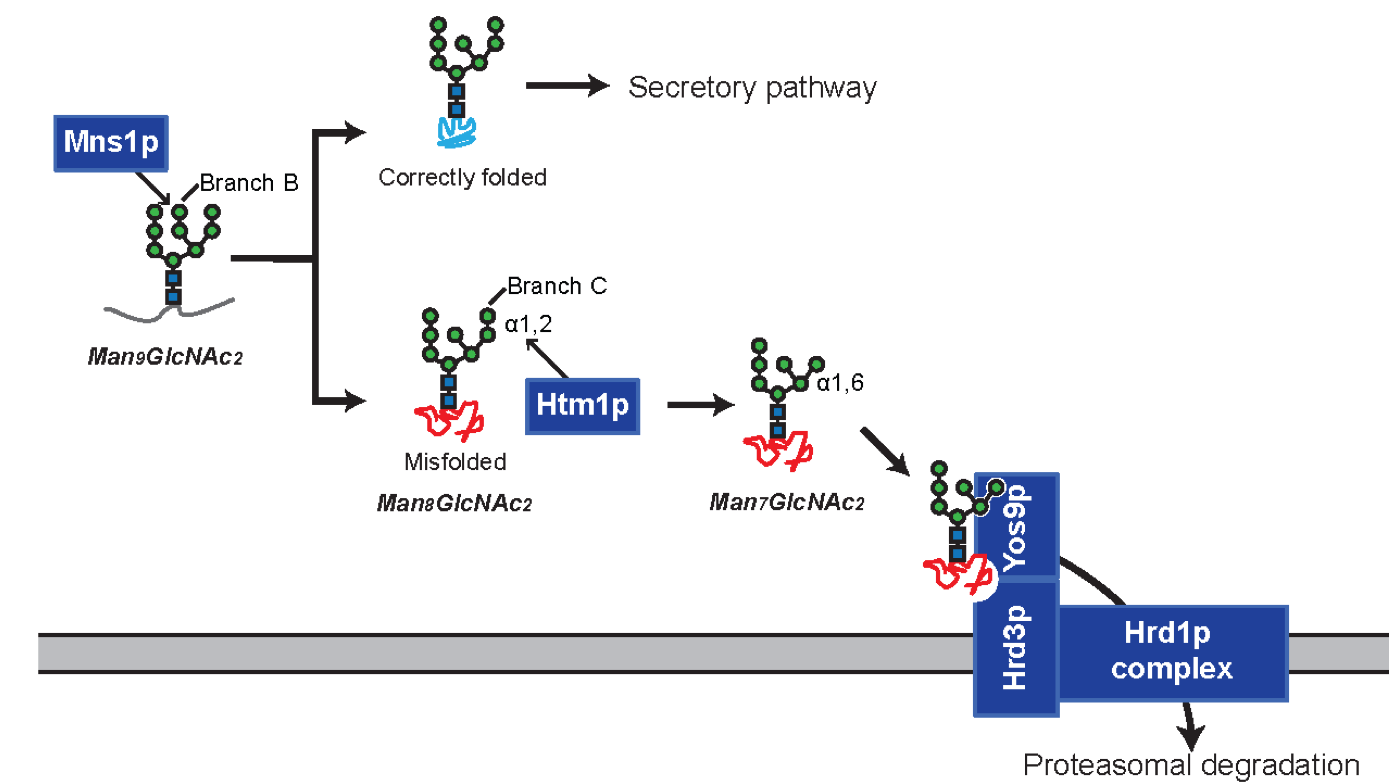
Liu YC, Fujimori DG, Weissman JS. Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc Natl Acad Sci USA 2016; 113(28):E4015-24 PMCID: PMC4948361 Full Text |
|
Pack LR, Yamamoto KR, Fujimori DG. Opposing Chromatin Signals Direct and Regulate the Activity of Lysine Demethylase 4C (KDM4C). J Biol Chem 2016 Mar 18; 291(12): 6060-70 PMCID: PMC4813556 Full Text |
|
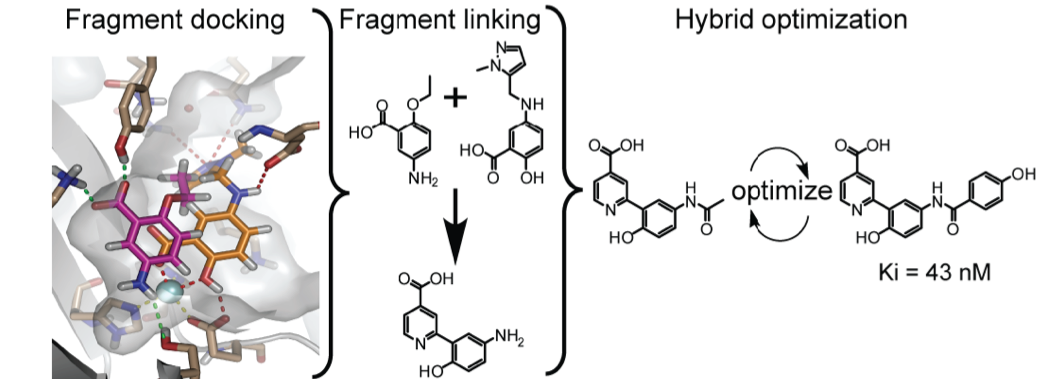
Korczynska M, Le DD, Younger N, Gregori-Puigjané E, Tumber A, Krojer T, Velupillai S, Gileadi C, Nowak RP, Iwasa E, Pollock SB, Ortiz Torres I, Oppermann U, Shoichet BK, Fujimori DG. Docking and Linking of Fragments To Discover Jumonji Histone Demethylase Inhibitors. J Med Chem 2016; 59(4):1 580-98
PMCID: PMC5080985 Full Text |
|
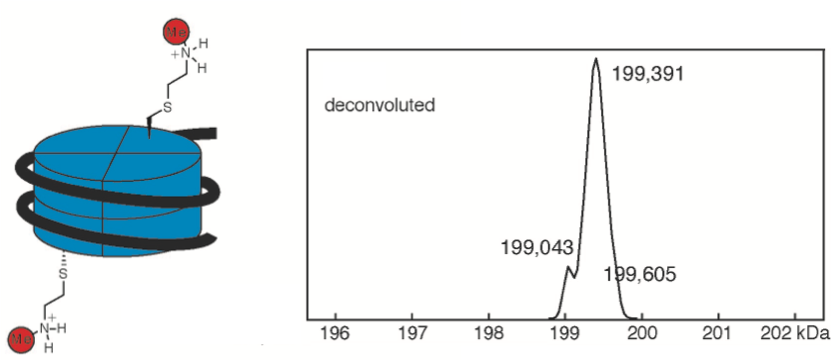
Lu J, Trnka, MJ, Roh SH, Robinson PJJ, Shiau C, Fujimori DG, Chiu W, Burlingame A, Guan G. Improved peak detection and deconvolution of native electrospray mass apectra from large protein complexes. J Am Soc Mass Spec 2015 Dec 26;26: 2141-51
PMCID: PMC5067139 Full Text |
|
Ortiz Torres I, Fujimori, DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol 2015 Nov 9;35: 68-75 PMCID: PMC4688207 Full Text |
|
Stojković, V, Fujimori DG. Radical SAM-mediated methylation of ribosomal RNA. Methods Enzymol 2015;560: 355-376 PMCID: PMC4530497 Full Text |
|
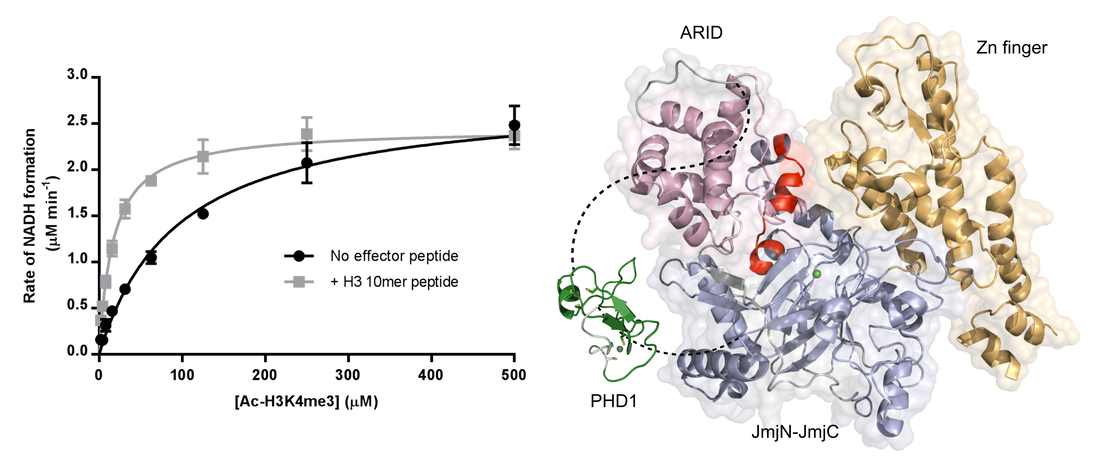
Ortiz Torres I, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat Commun 2015 Nov 17;6: 6204 PMCID: PMC5080983 Full Text |
|
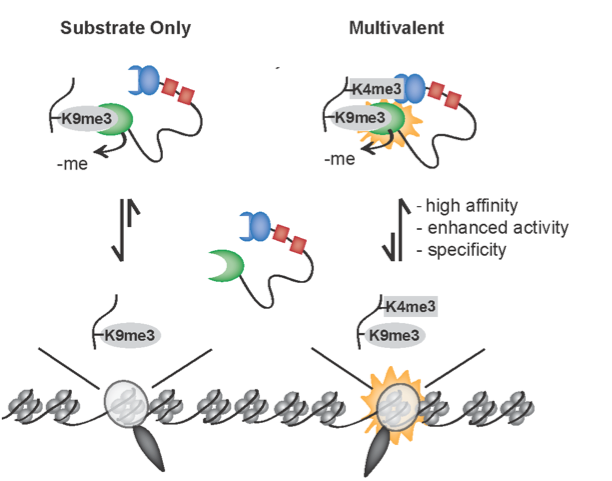
Dumesic PA, Homer CM, Moresco JJ, Pack LR, Coyle SM, Strahl BD, Fujimori DG, Yates III JR, Madhani HD. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 2015 Jan 15;160(1-2): 204-218 PMCID: PMC4303595 Full Text |
|
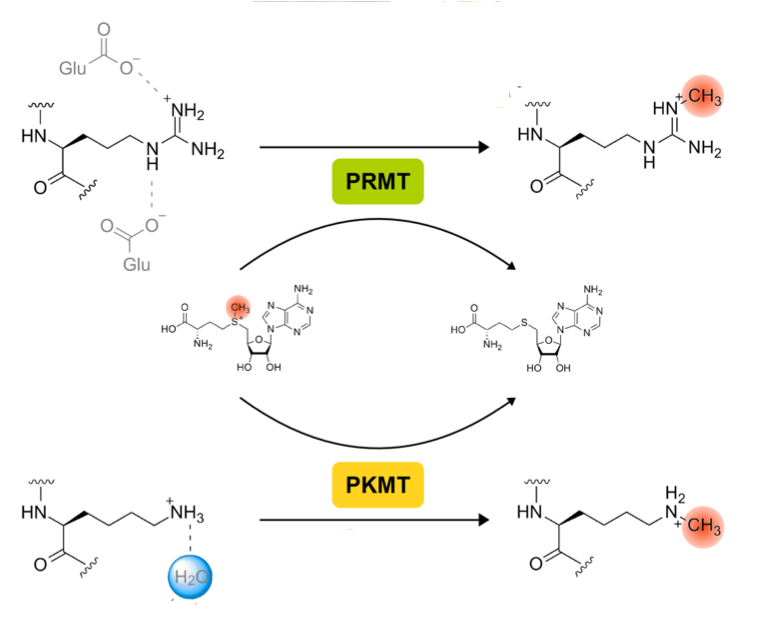
Fujimori DG. Radical SAM-mediated methylation reactions. Curr Opin Chem Biol 2013 Aug; 17: 597-604 (review) PMCID: PMC3799849 Full Text |
|
Shiau C, Trnka MJ, Bozicevic A, Ortiz Torres I, Al-Sady B, Burlingame AL, Narlikar GJ, Fujimori DG. Reconstitution of nucleosome demethylation and catalytic properties of a jumonji histone demethylase. Chem Biol 2013 Apr 18;20: 494-99 PMCID: PMC3704229 Full Text |
|
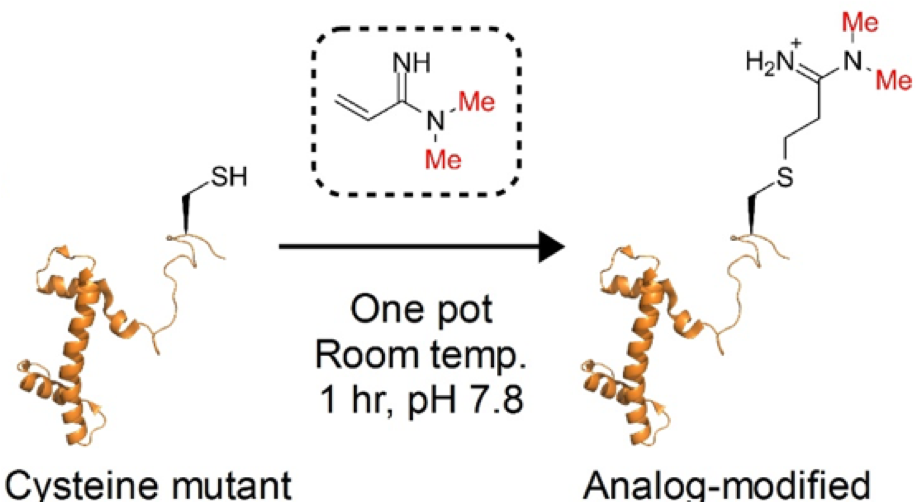
Le DD, Cortesi AT, Myers SA, Burlingame AL, Fujimori DG. Site-specific and regiospecific installation of methylarginine analogues into recombinant histones and insights into effector protein binding. J Am Chem Soc 2013 Feb 27;135: 2879-2882 PMCID: PMC4260808 Full Text |
|
Le DD, Fujimori DG. Protein and nucleic acid methylating enzymes: mechanisms and regulation. Curr Opin Chem Biol 2012 Dec;16: 507-515 PMCID: PMC3545634 Full Text |
|
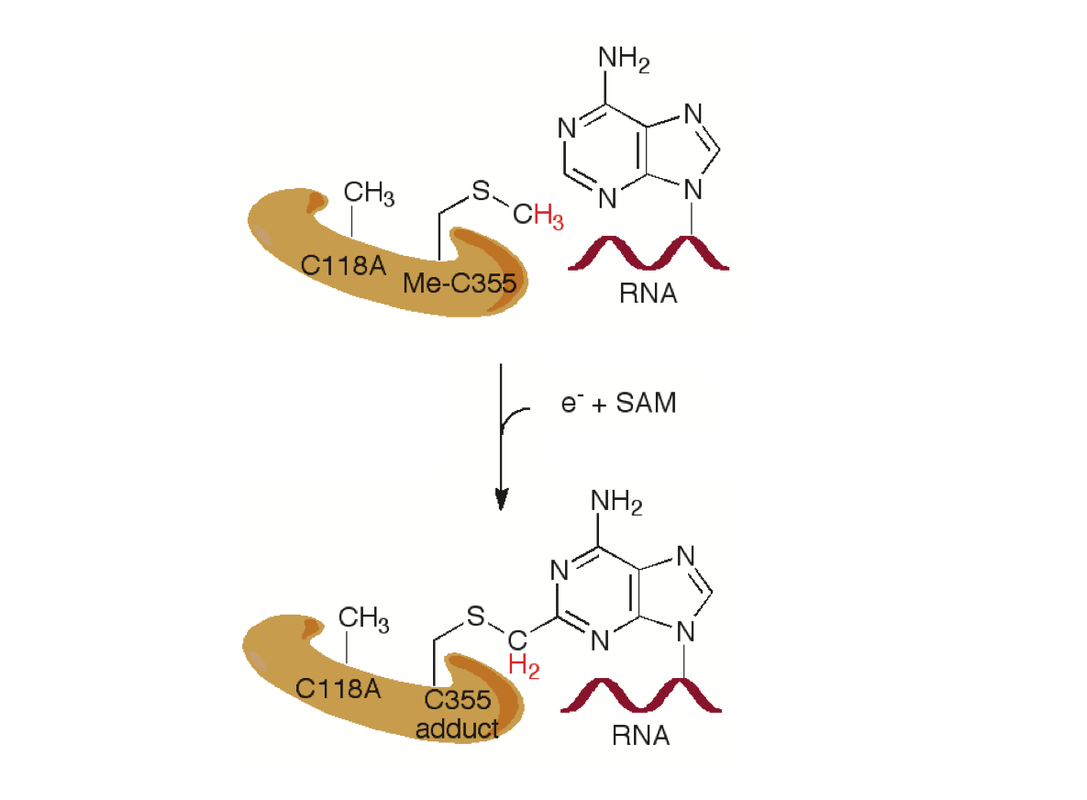
McCusker KP, Medzihradszky KF, Shiver AL, Nichols RJ, Yan F, Maltby DA, Gross CA, Fujimori DG. Covalent intermediate in the catalytic mechanism of the radical S-adenosyl-l-methionine methyl synthase RlmN trapped by mutagenesis. J Am Chem Soc 2012 Oct 31;134: 18074-81 PMCID: PMC3499099 Full Text |
|
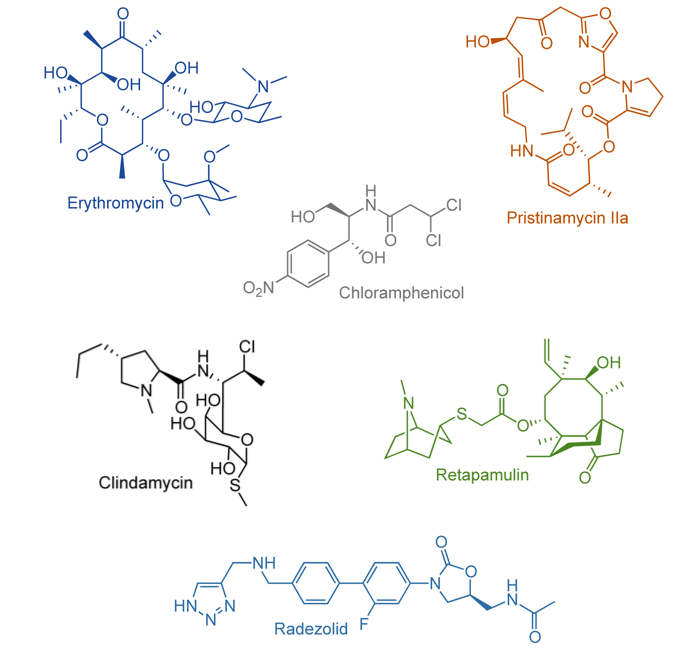
McCusker KP, Fujimori DG. The chemistry of peptidyltransferase center-targeted antibiotics: enzymatic resistance mechanisms and approaches to countering resistance, ACS Chem Biol 2012 Jan 20;7: 64-72 PMCID: PMC3499096 Full Text |
|
Yan, F, Fujimori, DG. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift, Proc Natl Acad Sci USA 2011 Mar 8;108: 3930-34 PMCID: PMC3054002 Full Text |
|
Yan F, LaMarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc 2010 Mar 24;132: 3953-64 PMCID: PMC2859901 Full Text |
Prior to UCSF (2000-2009)
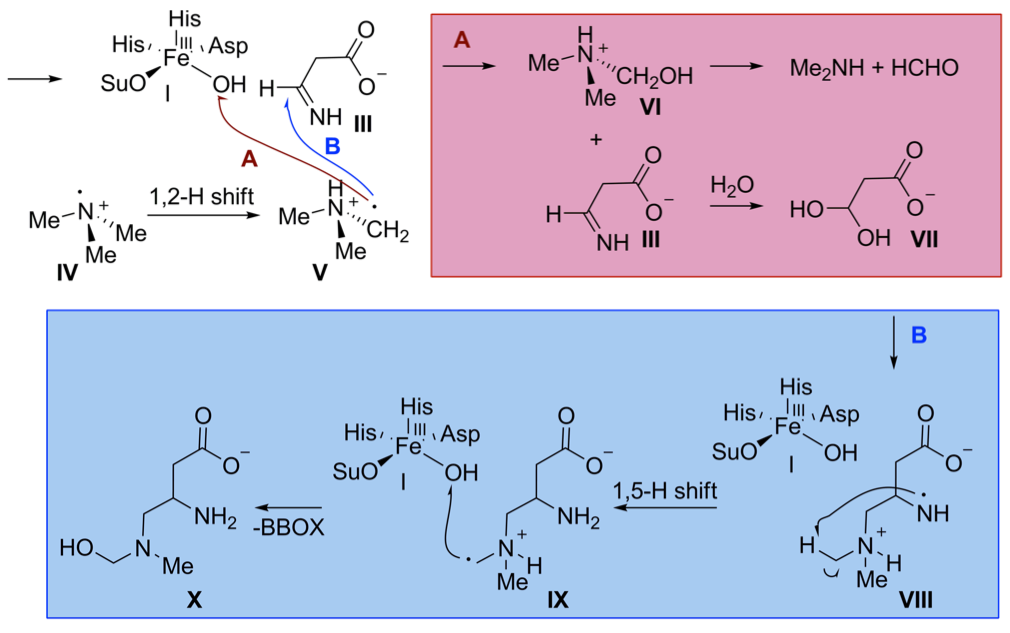
Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural analysis of an open site conformation of nonheme iron halogenase CytC3. J Am Chem Soc 131: 4872-9, 2009
PMCID: PMC2663892
Abstract
Full Text
Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural products biosynthesis. Chem Biol 15: 99-109, 2008 (review)
PMID: 18291314
Abstract
Full Text
Neidig ML, Brown CD, Light K, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM Jr, Krebs C, Walsh CT, Solomon EI. CD and MCD of CytC3 and taurine dioxygenase: role of facial triad in α-KG-dependent dioxygenases. J Am Chem Soc 129: 14224-31, 2007
PMCID: PMC2525739
Abstract
Full Text
Fujimori DG, Walsh CT. What’s new in enzymatic halogenations. Curr Opin Chem Biol 11: 553-60, 2007 (review)
PMCID: PMC2151916
Abstract
Full Text
Fujimori DG, Barr EW, Matthews ML, Koch GM, Yonce JR, Walsh CT, Bollinger Jr JM, Krebs C, Riggs-Gelasco PJ. Spectroscopic evidence for a high-spin Br-Fe(IV)-oxo intermediate in the α-ketoglutarate-dependent halogenase CytC3 from streptomyces. J Am Chem Soc 129: 13408-9, 2007
PMID: 17939667
Abstract
Full Text
Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci USA 104: 16498-503, 2007
PMCID: PMC2034236
Abstract
Full Text
Krebs C, Fujimori DG, Walsh CT, Bollinger Jr JM. Non-heme Fe(IV)-oxo intermediates. Acc Chem Res 40: 484-92, 2007 (review)
PMID: 17542550
Abstract
Full Text
Galonić DP, Barr EW, Walsh CT, Bollinger Jr JM, Krebs C. Two Interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nature Chem Biol 3: 113-6, 2007
PMID: 17220900
Abstract
Full Text
Kelly WL, Boyne II MT, Yeh E, Vosburg DA, Galonić DP, Kelleher NL, Walsh CT. Characterization of the aminocarboxycyclopropane-forming enzyme CmaC. Biochemistry 46: 359-68, 2007
PMID: 17209546
Abstract
Full Text
Galonić DP, Gin DY. Chemical glycosylation in the synthesis of glycoconjugate antitumor vaccines. Nature 446: 1000-7, 2007
PMCID: PMC2631661
Abstract
Full Text
Ueki M, Galonić DP, Vaillancourt FH, Garneau-Tsodikova S, Yeh E, Vosburg DA, Schroeder FC, Osada H, Walsh CT. Enzymatic generation of the antimetabolite γ,γ-dichloroaminobutyrate by NRPS and mononuclear iron halogenase action in a streptomycete. Chem Biol 13: 1183-91, 2006
PMID: 17114000
Abstract
Full Text
Galonić DP, Vaillancourt FH, Walsh CT. Halogenation of unactivated carbon centers in natural product biosynthesis: Trichlorination of leucine during barbamide biosynthesis. J Am Chem Soc 128: 3900-1, 2006
PMID: 16551084
Abstract
Full Text
Ide ND, Galonić DP, van der Donk WA, Gin DY. Conjugation of selenols with aziridine-2-carboxylic acid-containing peptides. Synlett 13: 2011-4, 2005
PMID: 15469231
Abstract
Full Text
Galonić DP, Ide ND, van der Donk WA, Gin DY. Aziridine-2-Carboxylic Acid Containing Peptides. Application to Solution- and Solid-Phase Convergent Site-Selective Peptide Modification. J Am Chem Soc 127: 7359-69, 2005
PMID: 15898784
Abstract
Full Text
Galonić DP, van der Donk WA, Gin DY. Convergent Site-Selective Ligation with Aziridine-2-Carboxylic Acid Containing Peptides. J Am Chem Soc 126: 12712-2, 2004
PMID: 15469231
Abstract
Full Text
Galonić DP, van der Donk WA, Gin DY. Oligosaccharide-Peptide Ligation of Glycosyl Thiolates with Dehydropeptides. Synthesis of S-Linked Mucin Glycopeptide Conjugates. Chem Eur J 9: 5997-6006, 2003
PMID: 14679512
Abstract
Full Text
Gieselman MD, Zhu Y, Zhou H, Galonić D, van der Donk WA. Selenocysteine Derivatives for Chemoselective Ligations. Chem Bio Chem 3: 709-16, 2002 (review)
PMID: 12203969
Abstract
Full Text
Lalić G, Galonić D, Matović R, Saičic RN. Model Study of Epothilone Synthesis: An Alternative Synthetic Approach to the C1-C7 fragment. J Serb Chem Soc 67: 221-7, 2002
Full Text
Lalić G, Petrovski Z, Galonić D, Matović R, Saičić RN. Alkylation of Carbonyl Compounds in the TiCl4-promoted Reaction of Trimethylsilyl Enol Ethers with Epoxides. Tetrahedron 57: 583-91, 2001
Full Text
Lalić G, Petrovski Z, Galonić D, Matović R, Saičić RN. Alkylation of Carbonyl Compounds in the TiCl4-promoted Reaction of Trimethylsilyl Enol Ethers with Ethylene Oxide. Tetrahedron Lett 41: 763-6, 2000
Full Text
PMCID: PMC2663892
Abstract
Full Text
Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural products biosynthesis. Chem Biol 15: 99-109, 2008 (review)
PMID: 18291314
Abstract
Full Text
Neidig ML, Brown CD, Light K, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM Jr, Krebs C, Walsh CT, Solomon EI. CD and MCD of CytC3 and taurine dioxygenase: role of facial triad in α-KG-dependent dioxygenases. J Am Chem Soc 129: 14224-31, 2007
PMCID: PMC2525739
Abstract
Full Text
Fujimori DG, Walsh CT. What’s new in enzymatic halogenations. Curr Opin Chem Biol 11: 553-60, 2007 (review)
PMCID: PMC2151916
Abstract
Full Text
Fujimori DG, Barr EW, Matthews ML, Koch GM, Yonce JR, Walsh CT, Bollinger Jr JM, Krebs C, Riggs-Gelasco PJ. Spectroscopic evidence for a high-spin Br-Fe(IV)-oxo intermediate in the α-ketoglutarate-dependent halogenase CytC3 from streptomyces. J Am Chem Soc 129: 13408-9, 2007
PMID: 17939667
Abstract
Full Text
Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci USA 104: 16498-503, 2007
PMCID: PMC2034236
Abstract
Full Text
Krebs C, Fujimori DG, Walsh CT, Bollinger Jr JM. Non-heme Fe(IV)-oxo intermediates. Acc Chem Res 40: 484-92, 2007 (review)
PMID: 17542550
Abstract
Full Text
Galonić DP, Barr EW, Walsh CT, Bollinger Jr JM, Krebs C. Two Interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nature Chem Biol 3: 113-6, 2007
PMID: 17220900
Abstract
Full Text
Kelly WL, Boyne II MT, Yeh E, Vosburg DA, Galonić DP, Kelleher NL, Walsh CT. Characterization of the aminocarboxycyclopropane-forming enzyme CmaC. Biochemistry 46: 359-68, 2007
PMID: 17209546
Abstract
Full Text
Galonić DP, Gin DY. Chemical glycosylation in the synthesis of glycoconjugate antitumor vaccines. Nature 446: 1000-7, 2007
PMCID: PMC2631661
Abstract
Full Text
Ueki M, Galonić DP, Vaillancourt FH, Garneau-Tsodikova S, Yeh E, Vosburg DA, Schroeder FC, Osada H, Walsh CT. Enzymatic generation of the antimetabolite γ,γ-dichloroaminobutyrate by NRPS and mononuclear iron halogenase action in a streptomycete. Chem Biol 13: 1183-91, 2006
PMID: 17114000
Abstract
Full Text
Galonić DP, Vaillancourt FH, Walsh CT. Halogenation of unactivated carbon centers in natural product biosynthesis: Trichlorination of leucine during barbamide biosynthesis. J Am Chem Soc 128: 3900-1, 2006
PMID: 16551084
Abstract
Full Text
Ide ND, Galonić DP, van der Donk WA, Gin DY. Conjugation of selenols with aziridine-2-carboxylic acid-containing peptides. Synlett 13: 2011-4, 2005
PMID: 15469231
Abstract
Full Text
Galonić DP, Ide ND, van der Donk WA, Gin DY. Aziridine-2-Carboxylic Acid Containing Peptides. Application to Solution- and Solid-Phase Convergent Site-Selective Peptide Modification. J Am Chem Soc 127: 7359-69, 2005
PMID: 15898784
Abstract
Full Text
Galonić DP, van der Donk WA, Gin DY. Convergent Site-Selective Ligation with Aziridine-2-Carboxylic Acid Containing Peptides. J Am Chem Soc 126: 12712-2, 2004
PMID: 15469231
Abstract
Full Text
Galonić DP, van der Donk WA, Gin DY. Oligosaccharide-Peptide Ligation of Glycosyl Thiolates with Dehydropeptides. Synthesis of S-Linked Mucin Glycopeptide Conjugates. Chem Eur J 9: 5997-6006, 2003
PMID: 14679512
Abstract
Full Text
Gieselman MD, Zhu Y, Zhou H, Galonić D, van der Donk WA. Selenocysteine Derivatives for Chemoselective Ligations. Chem Bio Chem 3: 709-16, 2002 (review)
PMID: 12203969
Abstract
Full Text
Lalić G, Galonić D, Matović R, Saičic RN. Model Study of Epothilone Synthesis: An Alternative Synthetic Approach to the C1-C7 fragment. J Serb Chem Soc 67: 221-7, 2002
Full Text
Lalić G, Petrovski Z, Galonić D, Matović R, Saičić RN. Alkylation of Carbonyl Compounds in the TiCl4-promoted Reaction of Trimethylsilyl Enol Ethers with Epoxides. Tetrahedron 57: 583-91, 2001
Full Text
Lalić G, Petrovski Z, Galonić D, Matović R, Saičić RN. Alkylation of Carbonyl Compounds in the TiCl4-promoted Reaction of Trimethylsilyl Enol Ethers with Ethylene Oxide. Tetrahedron Lett 41: 763-6, 2000
Full Text
Proudly powered by Weebly